python科学绘图
风物长宜放眼量,小不忍则乱大谋。(与君共勉)
终于将预测脂水分配系数的工具投出去了,腾出部分时间来进行相关的总结了。(我把Manuscript和Supporting information也一起放在这篇博客的后面,感兴趣的朋友可以取用,如果能引用就更加欢迎了!)
上次做完组会报告,隔壁实验室的师弟问我要了ppt说是想学习下我的作图,心中暗想自己的作图居然能得到部分认可,本着授之以鱼,不如授之以渔的观念,我干脆把我作图用到的脚本也全都online吧,一来可以做个科研记录,二来如果对志同道合的朋友有所帮助岂不美哉!
因为这篇工作99%的工作都是在python的框架下完成的,利用matplotlib工具包绘制图片远比利用gnuplot要方便地多,所以后续呈现的图也绝大多数都是利用python实现的。
本教程中用到的相关库的版本如下,大家也可以不必局限于这个版本,本教程中的图都是正文和SI中出现的子图,整个图片的整合可以利用各自喜欢的软件来进行,我比较倾向于利用power point来进行组合,大家可以用Photoshop等工具来进行。
Requirement
- Python
3.8 - numpy
1.21.1 - pandas
1.3.1 - matplotlib
3.4.2 - rdkit
2021.03.5
Figures
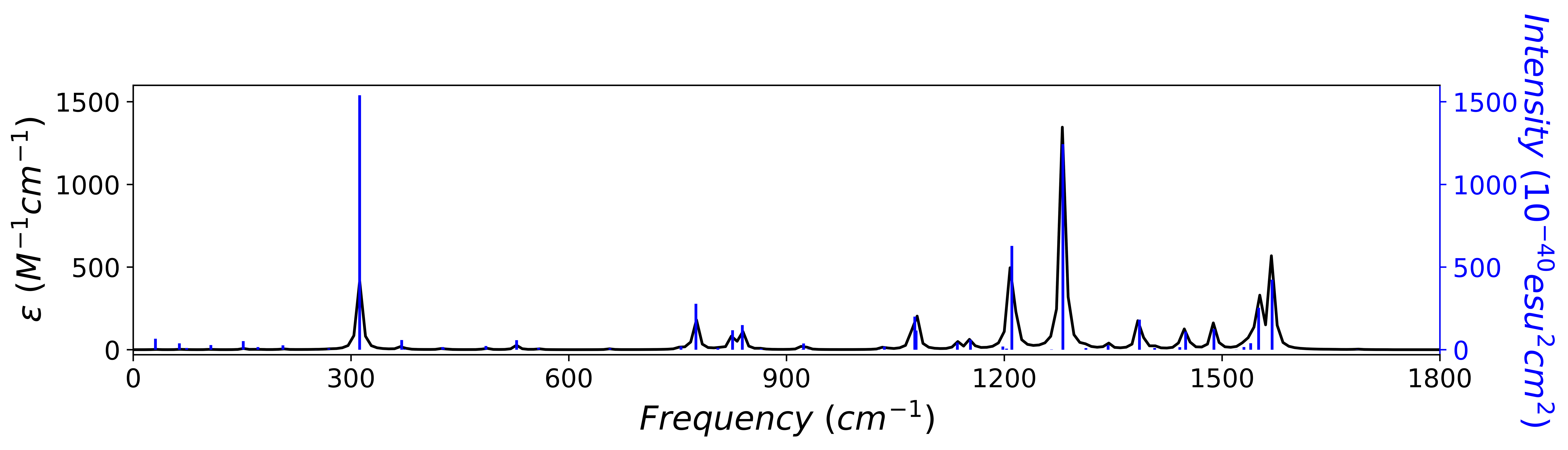
红外光谱图绘制
highlighted points:
- 绘制双Y轴并对两个轴着不同的颜色;
- 绘制离散的竖线,利用
vline来进行绘制
#!/bin/python
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# gaussian文件保存的红外光谱数据
filename = 'example_ir.txt'
# 读取文件中对应的数据
# 孤立的峰值数据
peak = []
# 通过展宽得到的连续的数据
broad = []
with open(filename,'r') as f:
while True:
line = f.readline()
if not line:
break
else:
if '# Peak information' in line:
line = f.readline()
for x in range(100000):
line = f.readline()
if '#' not in line:
break
else:
peak.append([float(line.split()[1]),float(line.split()[2])])
with open(filename,'r') as f:
while True:
line = f.readline()
if not line:
break
else:
if '# Spectra' in line:
line = f.readline()
for x in range(1000000):
line = f.readline()
if len(line.strip()) == 0:
break
else:
broad.append([float(line.split()[0]),float(line.split()[1]),float(line.split()[2])])
#print(np.array(peak))
#print(np.array(broad))
# 初始化画布
fig, ax = plt.subplots(figsize=(12,8))
peak = np.array(peak)
broad = np.array(broad)
#print(peak.shape[0])
ax.plot(broad[:,0],broad[:,1],color='black',linestyle='-')
ax2 = ax.twinx()
for x in np.arange(peak.shape[0]):
ax2.vlines(peak[x][0],0,peak[x][1],color='blue',linestyle='-')
ax2.set_ylim(-30,1600)
ax2.yaxis.set_ticks(np.arange(0,1500.1,500))
ax2.yaxis.set_tick_params(labelsize=14, color='blue')
ax2.spines['right'].set_color('blue')
ax2.xaxis.label.set_color('blue')
ax2.tick_params(axis='y',colors='blue')
ax2.set_ylabel(r'$Intensity~(10^{-40} esu^2 cm^2)$',fontsize=18,fontfamily='sans-serif',fontstyle='italic',color='blue',rotation=-90,labelpad=20)
ax.set_xlabel(r'Frequency $(cm^{-1})$',fontsize=18,fontfamily='sans-serif',fontstyle='italic')
ax.set_ylabel(r'$\epsilon~(M^{-1}cm^{-1})$',fontsize=18,fontfamily='sans-serif',fontstyle='italic')
ax.set_xlim(0,1800)
ax.set_ylim(-30,1600)
ax.xaxis.set_tick_params(labelsize=14)
ax.yaxis.set_tick_params(labelsize=14)
ax.xaxis.set_ticks(np.arange(0,1800.1,300))
ax.yaxis.set_ticks(np.arange(0,1500.1,500))
plt.tight_layout()
plt.savefig('examplt_ir_broaden.png',dpi=1000,bbox_inches='tight')
plt.show()
静电势的绘制(非python绘图)
- 首先利用
gaussian产生对应的density.cube和potential.cube文件,具体命令如下:formchk *.chk *.fchk cubegen 0 density *.fchk density.cube 0 h & cubegeb 0 potential *.fchk potential.cube 0 h & # 相关的命令解释我就不做介绍了,大家有兴趣的可以去gaussian官网上查询! - 利用sob老师给出的
ESPiso.vmd脚本产生对应的静电势图 (致谢师妹付路路!)#This script is used to draw ESP colored molecular vdW surface (rho=0.001) #density1.cub, ESP1.cub, density2.cub, ESP2.cub ... should be presented in current folder color scale method BWR color Display Background white axes location Off display depthcue off display rendermode GLSL light 2 on light 3 on material change transmode EdgyGlass 1.000000 #The maximum number of systems to be loaded set nsystem 1 #Lower and upper limit of color scale of ESP (a.u.) set colorlow -0.03 set colorhigh 0.03 for {set i 1} {$i<=$nsystem} {incr i} { set id [expr $i-1] mol new density.cube mol addfile potential$i.cube # this should be replaced with your defined filename mol modstyle 0 $id CPK 1.000000 0.300000 22.000000 22.000000 mol addrep $id mol modstyle 1 $id Isosurface 0.001000 0 0 0 1 1 mol modmaterial 1 $id EdgyGlass mol modcolor 1 $id Volume 1 mol scaleminmax $id 1 $colorlow $colorhigh }
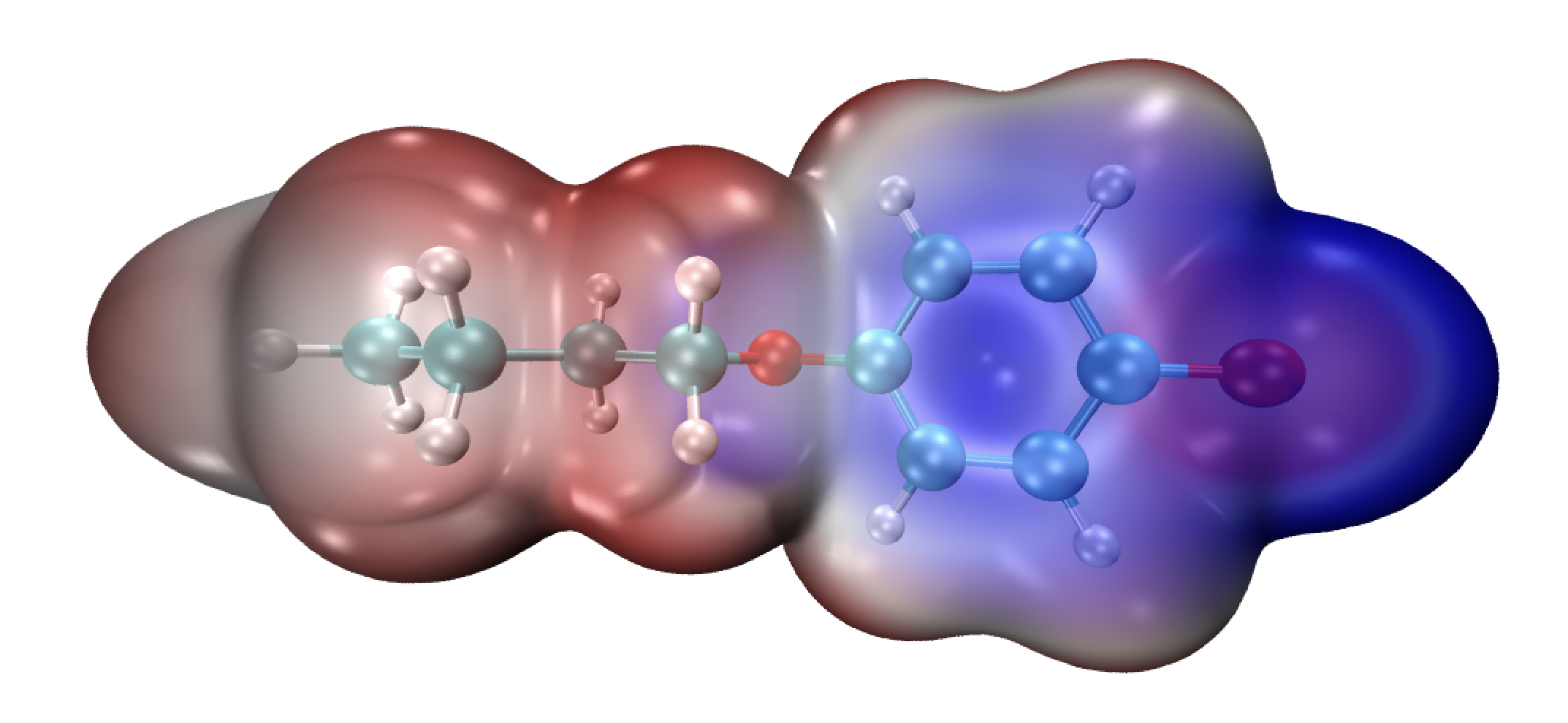
script:ESPiso.vmd
绘制gaussian计算得到的振动模式 (非python绘图)
同样是结合VMD食用,致谢付路路师妹!
if you have any questions, please ref http://sobereva.com/567
Usage:
source GauNorm.tcl
norm $imode $scalfac $rad $color
- imode: the index of the mode
- sclfac: length of the vector
- rad: radius of the arrow
- colors: default set to be yellow
scripts:GauNorm.tcl, drawarrow.tcl
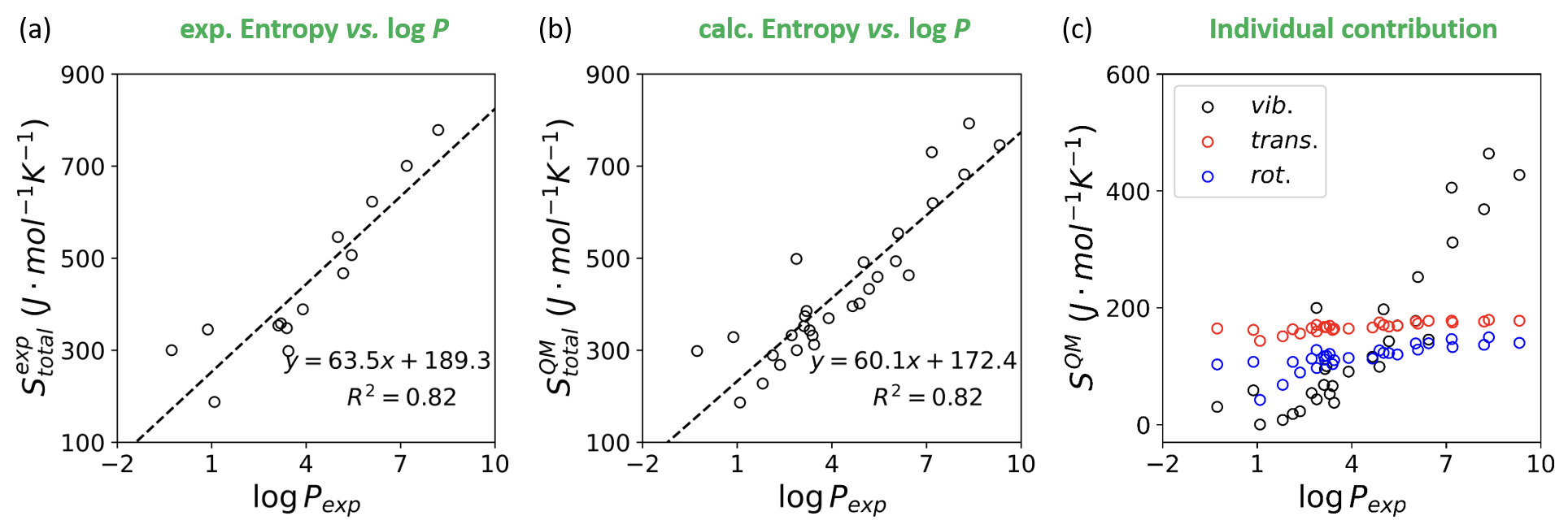
绘制中空的散点图并拟合一条直线
highlight:
- linear fitting curves
- scatters without filling colors
# 首先定义计算R2的函数
def r2_plot(x, y):
mod = LinearRegression()
mod.fit(np.array(x).reshape(-1,1), np.array(y).reshape(-1,1))
b = mod.intercept_
a = mod.coef_
score = mod.score(np.array(x).reshape(-1,1), np.array(y).reshape(-1,1))
return a[0],b,score
r0,r1,r2 = r2_plot(dataset_exp['logp'],dataset_exp['entropy-exp'])
r0,r1,r2
#绘制相关的图片
fig, ax = plt.subplots(figsize=(4,4))
ax.scatter(dataset_exp['logp'],dataset_exp['entropy-exp'],color='none',marker='o',edgecolor='black')
ax.plot(np.arange(-2,10.1,1), a1*np.arange(-2,10.1,1)+a0, linestyle='--',color='black',label='fitted line')
ax.text(3.3,260,'$y={:.1f}x+{:.1f}$'.format(r0[0],r1[0]),size=14,color='black')
ax.text(5.3,180,r'$R^2={:.2f}$'.format(r2),size=14,color='black')
ax.xaxis.set_tick_params(labelsize=14)
ax.yaxis.set_tick_params(labelsize=14)
ax.set_xlim(-2,10)
ax.set_ylim(100,800)
ax.xaxis.set_ticks(np.arange(-2,10.1,3))
ax.yaxis.set_ticks(np.arange(100,900.1,200))
ax.set_xlabel(r'$\log P_{exp}$',fontsize=18,fontfamily='sans-serif',fontstyle='italic')
ax.set_ylabel(r'$S^{exp}_{total}~(J\cdot mol^{-1}K^{-1})$',fontsize=18,fontfamily='sans-serif',fontstyle='italic')
#ax.legend(fontsize=14)
plt.savefig('Entropy_vs_logP/entropy_vs_logp.png',dpi=1000,bbox_inches='tight')
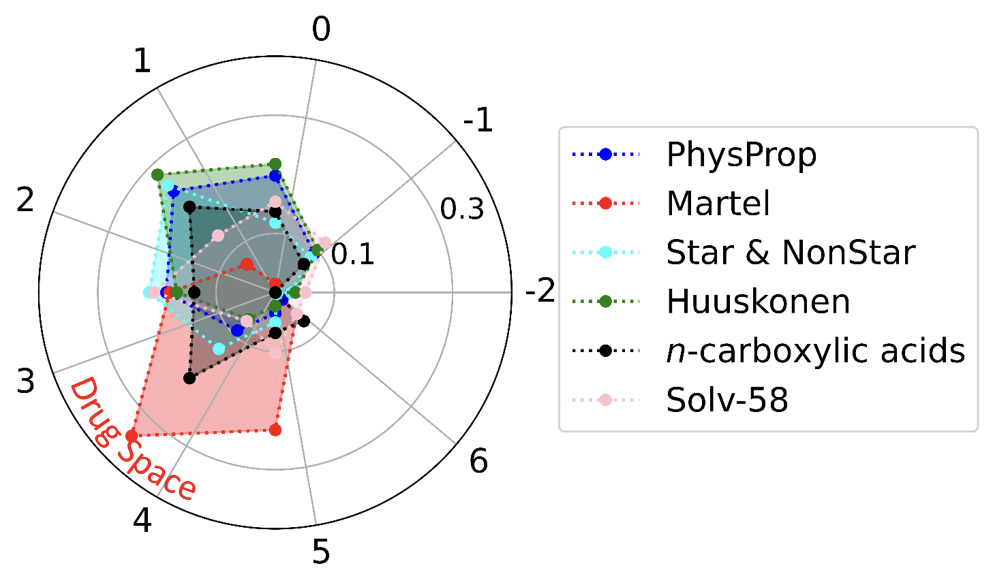
绘制雷达图
highlights:
- radar plot
# importing package
from logp.io.elem_filter import filt_elems # function written by myself
from logp.zq.utils.element_count import get_element
import os, warnings
import pandas as pd
warnings.filterwarnings('ignore')
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# data processing
# Specify the element you want to fileter
# elems_specified = ['C','H','O','N']
# load the dataset we collected
dataset = 'logp/DATASETS/DATASETS_TOTAL.xlsx'
dataset_martel = pd.read_excel(dataset,sheet_name='Martel')
dataset_physprop = pd.read_excel(dataset,sheet_name='PhysProp')
f_star = 'logp/DATASETS/star.xls'
f_nonstar = 'logp/DATASETS/nonstar.xls'
dataset_star = pd.read_excel(f_star,sheet_name='star.xls')
dataset_nonstar = pd.read_excel(f_nonstar,sheet_name='nonstar.xls')
#dataset_filt = filt_elems(dataset_martel,elems_specified)
#dataset_filt = pd.DataFrame(data=dataset_filt)
f_acids = 'logp/DATASETS/n-carboxylic_acids.xlsx'
dataset_acids = pd.read_excel(f_acids)
f_solv_58 = 'logp/DATASETS/Solv-58/Solv-58.xlsx'
dataset_solv = pd.read_excel(f_solv_58)
# count element
elem_martel = get_element(dataset_martel['SMILES'])
elem_physprop = get_element(dataset_physprop['SMILES'])
elem_star = get_element(dataset_star['SMILES'])
elem_nonstar = get_element(dataset_nonstar['SMILES'])
elem_acids = get_element(dataset_acids['smile'])
elem_solv = get_element(dataset_solv['smiles'])
# summary the total element of the molecules we studied
df_martel = pd.DataFrame(elem_martel)
df_martel = df_martel.set_index(['name'])
df_martel.loc['col_sum'] = df_martel.apply(lambda x: x.sum())
df_physprop = pd.DataFrame(elem_physprop)
df_physprop = df_physprop.set_index(['name'])
df_physprop.loc['col_sum'] = df_physprop.apply(lambda x: x.sum())
df_star = pd.DataFrame(elem_star)
df_star = df_star.set_index(['name'])
df_star.loc['col_sum'] = df_star.apply(lambda x: x.sum())
df_nonstar = pd.DataFrame(elem_nonstar)
df_nonstar = df_nonstar.set_index(['name'])
df_nonstar.loc['col_sum'] = df_nonstar.apply(lambda x: x.sum())
df_acids = pd.DataFrame(elem_acids)
df_acids = df_acids.set_index(['name'])
df_acids.loc['col_sum'] = df_acids.apply(lambda x: x.sum())
df_solv = pd.DataFrame(elem_solv)
df_solv = df_solv.set_index(['name'])
df_solv.loc['col_sum'] = df_solv.apply(lambda x: x.sum())
# get the LogP values
logp_physprop = dataset_physprop['logp'].values
logp_martel = dataset_martel['logp'].values
logp_star = dataset_star['logPow {measured}'].values
logp_nonstar = dataset_nonstar['logPow {measured}'].values
logp_acids = dataset_acids['logp_exp'].values
logp_solv = dataset_solv['logp'].values
# set the resolution of the histogram
bins =8
ax2 = plt.subplot(projection='polar')
#ax1 = plt.subplot2grid((1,2), (0,1), rowspan=2, colspan=2,polar=True)
theta = np.linspace(0, 2*np.pi, bins, endpoint=False)
theta = np.concatenate((theta,[theta[0]]))
print(type(logp_physprop))
# get the histogram info
hist_physprop, bin_edges_physprop = np.histogram(logp_physprop,bins=bins,range=(-2,7), density=True)
hist_martel, bin_edges_martel = np.histogram(logp_martel,bins=bins,range=(-2,7),density=True)
hist_star, bin_edges_star = np.histogram(logp_star,bins=bins,range=(-2,7),density=True)
hist_huuskonen, bin_edges_huuskonen = np.histogram(logp_nonstar,bins=bins,range=(-2,7),density=True)
hist_acids, bin_edges_acids = np.histogram(logp_acids,bins=bins, range=(-2,7),density=True)
hist_solv, bin_edges_solv = np.histogram(logp_solv, bins=bins, range=(-2,7),density=True)
hist_physprop = np.concatenate((hist_physprop,[hist_physprop[0]]))
hist_martel = np.concatenate((hist_martel, [hist_martel[0]]))
hist_star = np.concatenate((hist_star, [hist_star[0]]))
hist_huuskonen = np.concatenate((hist_huuskonen, [hist_huuskonen[0]]))
hist_acids = np.concatenate((hist_acids, [hist_acids[0]]))
hist_solv = np.concatenate((hist_solv, [hist_solv[0]]))
ax2.set_thetagrids(np.arange(0.0, 360.0, 40),np.arange(-2,7,1))
ax2.xaxis.set_tick_params(labelsize=16)
ax2.yaxis.set_tick_params(labelsize=14)
ax2.set_rlim(0.,0.4)
ax2.set_rgrids(np.arange(0.1,0.41,0.2))
ax2.plot(theta,hist_physprop,'bo:',ms=5,label='PhysProp')
ax2.fill(theta,hist_physprop,color='b',alpha=0.3)
ax2.plot(theta,hist_martel,'ro:',ms=5,label='Martel')
ax2.fill(theta,hist_martel,color='r',alpha=0.3)
ax2.plot(theta,hist_star,color='cyan',marker='o',ls=':',ms=5,label='Star & NonStar')
ax2.fill(theta,hist_star,color='cyan',alpha=0.3)
ax2.plot(theta,hist_huuskonen,color='green',marker='o',ls=':',ms=5,label='Huuskonen')
ax2.fill(theta,hist_huuskonen,color='green',alpha=0.3)
ax2.plot(theta,hist_acids,color='black',marker='o',ls=':',ms=5,label=r'$n$-carboxylic acids')
ax2.fill(theta,hist_acids,color='black',alpha=0.3)
ax2.plot(theta,hist_solv,color='pink',marker='o',ls=':',ms=5,label=r'Solv-58')
ax2.fill(theta,hist_solv,color='pink',alpha=0.3)
plt.legend(bbox_to_anchor=(1.1, 0.85), loc=2, borderaxespad=0,fontsize=16)
plt.tight_layout()
plt.savefig('elem_counts/6_datasets_radar_logp_distribution.png',dpi=1000,bbox_inches='tight')
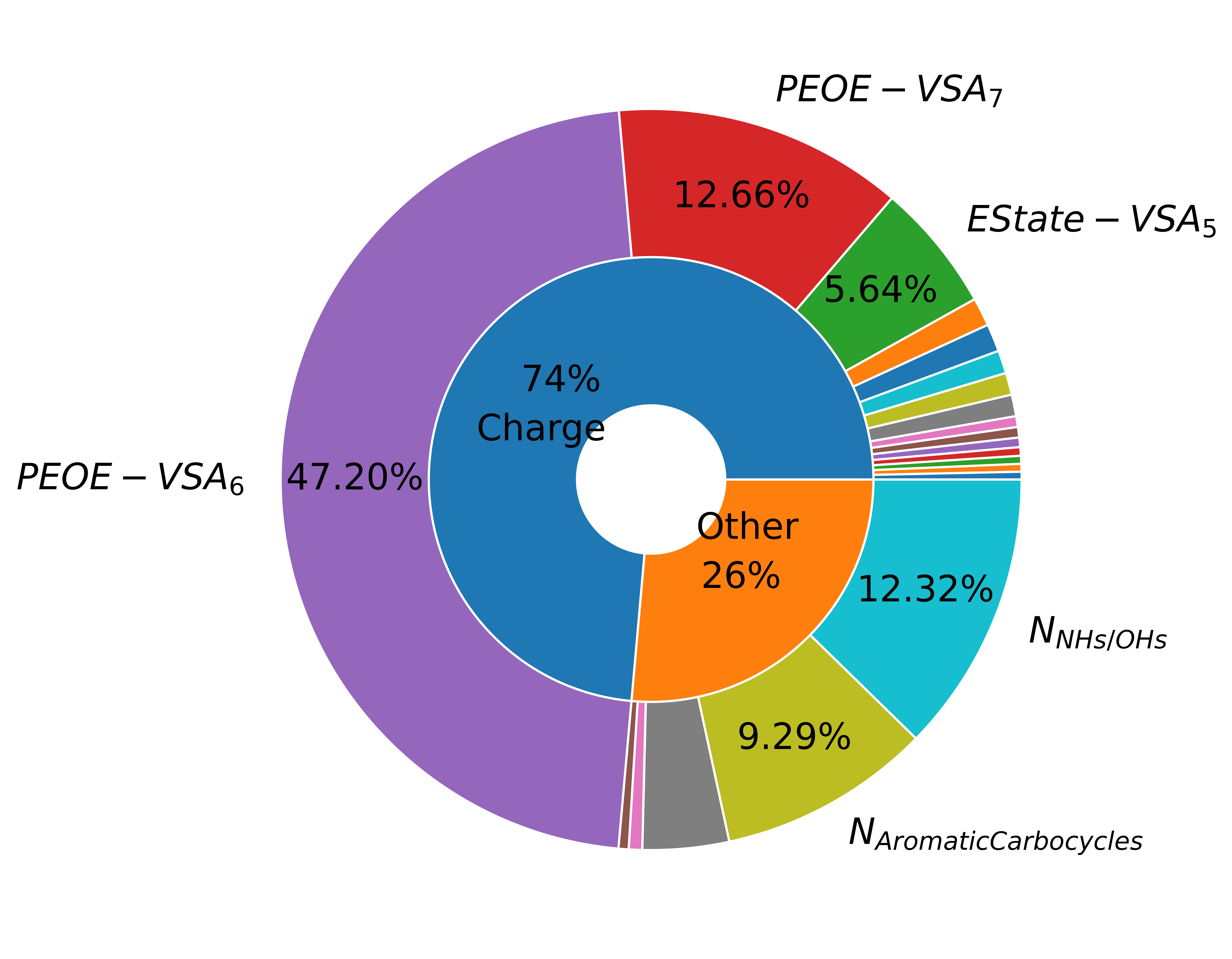
描述符重要性饼状图
highlights:
- 饼状图;
- 重要性按照比重排序;
- 重要性按照比重进行了分类
# importing the package
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# loading the data
pie_mdi = pd.read_csv('rf_mdi_importance.csv')
pie_mdi.head(5)
mdi_charge_sum = pie_mdi[pie_mdi['classify']=='charge']['pop'].sum()
mdi_other_sum = pie_mdi[pie_mdi['classify']=='non-charge']['pop'].sum()
# sorting the values so as to plot more beautifully
pie_mdi_sorted = pie_mdi.sort_values(by=['classify','pop'])
# initial the setting
fig, ax = plt.subplots(figsize=(10,6),subplot_kw=dict(aspect='equal'))
def my_autopct(pct):
return ('%.2f%%' %pct) if pct > 4 else ''
labels = [n if v > pie_mdi_sorted['pop'].sum() *0.04 else ''
for n,v in zip(pie_mdi_sorted['name'],pie_mdi_sorted['pop'])]
ax.pie(pie_mdi_sorted['pop'],labels=labels,
autopct=my_autopct,pctdistance=0.8,
radius=1, wedgeprops=dict(width=0.4, edgecolor='w'),textprops={'fontsize':16})
ax.pie([mdi_charge_sum,mdi_other_sum],labels=['Charge','Other'],
labeldistance=0.3,
autopct='%1.0f%%',pctdistance=0.6,
radius=1-0.4, wedgeprops=dict(width=0.4,edgecolor='w'),textprops={'fontsize':16})
plt.tight_layout()
plt.savefig('rf_feature_importance_pie.png',dpi=1000,bbox_inches='tight')
data: rf_mdi_importance.csv
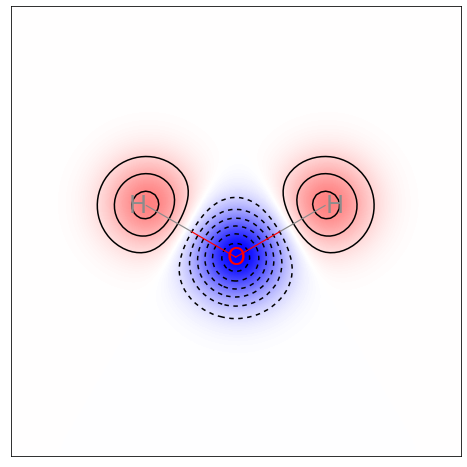
将原子的权重map到分子的二维结构上
highlights:
- 背景设成白色;
- 添加边框;
- 抹去XY相关信息
#from logp.sf_behler.sf import get_charge
#from logp.sf_behler.g3D import XYZ_dict
#from logp.sf_behler.sf import sum_G1
#from logp.sf_behler.sf import sum_G2
# 上述都是自己编写的函数,直接注释掉
from rdkit import Chem
from rdkit.Chem.Draw import SimilarityMaps
import matplotlib.pyplot as plt
# 获得水的原子电荷,这里RDKit自带的是Gasteiger partial charge
smile = 'O'
mol = Chem.MolFromSmiles(smile)
mH = Chem.AddHs(mol) #需要显式地表示H原子
charge = get_charge(mH)
fig1 = SimilarityMaps.GetSimilarityMapFromWeights(mH, charge, colorMap='jet',contourLines=10,scale=-1)
plt.axis('on') # 显示边框
plt.xticks([])
plt.yticks([])
fig1.savefig('SF_figures/water_q.png',bbox_inches='tight') #如果是在jupyter中运行的话一定要加上关键词bbox_inches,不然图片显示不完全
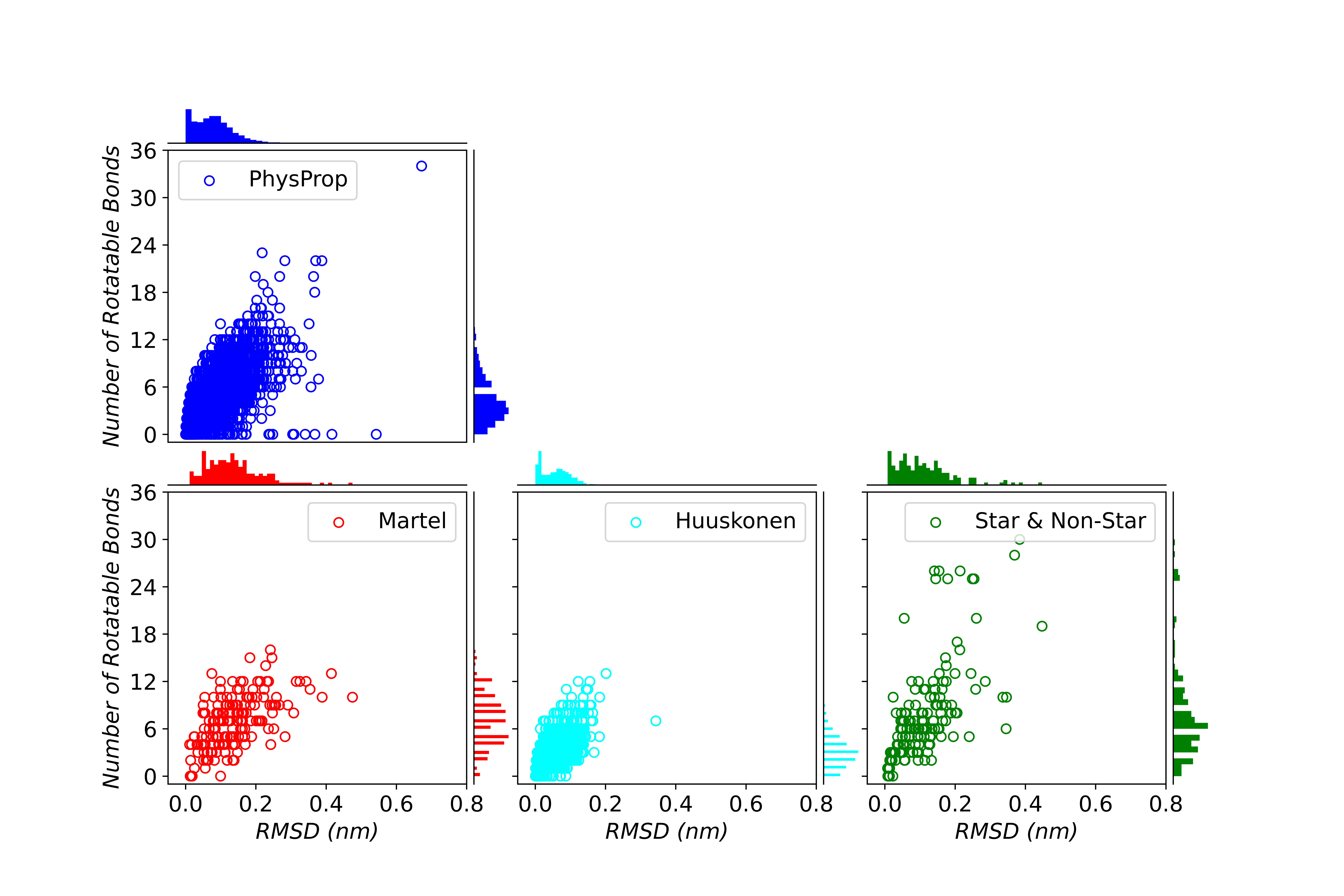
散点图+统计分布图在侧边
highlights:
- 多图绘制;
- 绘制散点图的同时,将分布图分布在两个坐标轴上;
fig = plt.figure(figsize=(12,8)) # 创建画布
#ax1 = plt.subplot(2,3,1)
Grid = plt.GridSpec(16,24,hspace=0.2,wspace=0.2) #确定哪些grid用来绘图,哪些区间是空缺状态
main_axes1 = fig.add_subplot(Grid[1:8,0:7],xticklabels=[])
y_hist1 = fig.add_subplot(Grid[1:8,7],xticklabels=[],sharey=main_axes1)
x_hist1 = fig.add_subplot(Grid[0,0:7],yticklabels=[],sharex=main_axes1)
main_axes1.scatter(physprop_rmsds,physprop_rot,color='none', marker='o', edgecolor='blue',label='PhysProp')
x_hist1.hist(physprop_rmsds,bins=40, histtype='stepfilled',orientation='vertical',color='blue',density=1.0)
y_hist1.hist(physprop_rot,bins=40, histtype='stepfilled',orientation='horizontal',color='blue',density=1.0)
x_hist1.tick_params(axis='x',labelbottom=False)
y_hist1.tick_params(axis='y',labelleft=False)
x_hist1.spines['right'].set_visible(False)
x_hist1.spines['left'].set_visible(False)
x_hist1.spines['top'].set_visible(False)
x_hist1.yaxis.set_ticks([])
x_hist1.xaxis.set_ticks([])
y_hist1.spines['right'].set_visible(False)
y_hist1.spines['bottom'].set_visible(False)
y_hist1.spines['top'].set_visible(False)
y_hist1.yaxis.set_ticks([])
y_hist1.xaxis.set_ticks([])
main_axes1.legend(fontsize=14)
main_axes1.set_xlim(-0.05,0.8)
main_axes1.set_ylim(-1,36)
#main_axes1.xaxis.set_ticks(np.arange(-0.0,0.81,0.2))
main_axes1.yaxis.set_ticks(np.arange(0,37,6))
main_axes1.xaxis.set_ticks([])
main_axes1.xaxis.set_tick_params(labelsize=14)
main_axes1.yaxis.set_tick_params(labelsize=14)
main_axes1.set_ylabel('Number of Rotatable Bonds',fontsize=14,fontfamily='sans-serif',fontstyle='italic')
main_axes2 = fig.add_subplot(Grid[9:,0:7])
y_hist2 = fig.add_subplot(Grid[9:,7],xticklabels=[],sharey=main_axes2)
x_hist2 = fig.add_subplot(Grid[8,0:7],yticklabels=[],sharex=main_axes2)
main_axes2.scatter(martel_rmsds,martel_rot,color='none', marker='o', edgecolor='red',label='Martel')
x_hist2.hist(martel_rmsds,bins=40, histtype='stepfilled',orientation='vertical',color='red',density=1.0)
y_hist2.hist(martel_rot,bins=40, histtype='stepfilled',orientation='horizontal',color='red',density=1.0)
x_hist2.tick_params(axis='x',labelbottom=False)
y_hist2.tick_params(axis='y',labelleft=False)
x_hist2.spines['right'].set_visible(False)
x_hist2.spines['left'].set_visible(False)
x_hist2.spines['top'].set_visible(False)
x_hist2.yaxis.set_ticks([])
x_hist2.xaxis.set_ticks([])
y_hist2.spines['right'].set_visible(False)
y_hist2.spines['bottom'].set_visible(False)
y_hist2.spines['top'].set_visible(False)
y_hist2.yaxis.set_ticks([])
y_hist2.xaxis.set_ticks([])
main_axes2.legend(fontsize=14)
main_axes2.set_xlim(-0.05,0.8)
main_axes2.set_ylim(-1,36)
main_axes2.xaxis.set_ticks(np.arange(-0.0,0.81,0.2))
main_axes2.yaxis.set_ticks(np.arange(0,37,6))
main_axes2.xaxis.set_tick_params(labelsize=14)
main_axes2.yaxis.set_tick_params(labelsize=14)
main_axes2.set_xlabel('RMSD (nm)',fontsize=14,fontfamily='sans-serif',fontstyle='italic')
main_axes2.set_ylabel('Number of Rotatable Bonds',fontsize=14,fontfamily='sans-serif',fontstyle='italic')
main_axes3 = fig.add_subplot(Grid[9:,8:15],yticklabels=[])
y_hist3 = fig.add_subplot(Grid[9:,15],xticklabels=[],sharey=main_axes3)
x_hist3 = fig.add_subplot(Grid[8,8:15],yticklabels=[],sharex=main_axes3)
main_axes3.scatter(huuskonen_rmsds,huuskonen_rot,color='none', marker='o', edgecolor='cyan',label='Huuskonen')
x_hist3.hist(huuskonen_rmsds,bins=40, histtype='stepfilled',orientation='vertical',color='cyan',density=1.0)
y_hist3.hist(huuskonen_rot,bins=40, histtype='stepfilled',orientation='horizontal',color='cyan',density=1.0)
x_hist3.tick_params(axis='x',labelbottom=False)
y_hist3.tick_params(axis='y',labelleft=False)
x_hist3.spines['right'].set_visible(False)
x_hist3.spines['left'].set_visible(False)
x_hist3.spines['top'].set_visible(False)
x_hist3.yaxis.set_ticks([])
x_hist3.xaxis.set_ticks([])
y_hist3.spines['right'].set_visible(False)
y_hist3.spines['bottom'].set_visible(False)
y_hist3.spines['top'].set_visible(False)
y_hist3.yaxis.set_ticks([])
y_hist3.xaxis.set_ticks([])
main_axes3.legend(fontsize=14)
main_axes3.set_xlim(-0.05,0.8)
main_axes3.set_ylim(-1,36)
main_axes3.xaxis.set_ticks(np.arange(-0.0,0.81,0.2))
main_axes3.yaxis.set_ticks(np.arange(0,37,6))
main_axes3.xaxis.set_tick_params(labelsize=14)
main_axes3.yaxis.set_tick_params(labelsize=14)
main_axes3.set_xlabel('RMSD (nm)',fontsize=14,fontfamily='sans-serif',fontstyle='italic')
main_axes4 = fig.add_subplot(Grid[9:,16:23],yticklabels=[])
y_hist4 = fig.add_subplot(Grid[9:,23],xticklabels=[],sharey=main_axes4)
x_hist4 = fig.add_subplot(Grid[8,16:23],yticklabels=[],sharex=main_axes4)
main_axes4.scatter(star_rmsds,star_rot,color='none', marker='o', edgecolor='green',label='Star & Non-Star')
x_hist4.hist(star_rmsds,bins=40, histtype='stepfilled',orientation='vertical',color='green',density=1.0)
y_hist4.hist(star_rot,bins=40, histtype='stepfilled',orientation='horizontal',color='green',density=1.0)
x_hist4.tick_params(axis='x',labelbottom=False)
y_hist4.tick_params(axis='y',labelleft=False)
x_hist4.spines['right'].set_visible(False)
x_hist4.spines['left'].set_visible(False)
x_hist4.spines['top'].set_visible(False)
x_hist4.yaxis.set_ticks([])
x_hist4.xaxis.set_ticks([])
y_hist4.spines['right'].set_visible(False)
y_hist4.spines['bottom'].set_visible(False)
y_hist4.spines['top'].set_visible(False)
y_hist4.yaxis.set_ticks([])
y_hist4.xaxis.set_ticks([])
main_axes4.legend(fontsize=14)
main_axes4.set_xlim(-0.05,0.8)
main_axes4.set_ylim(-1,36)
main_axes4.xaxis.set_ticks(np.arange(-0.0,0.81,0.2))
main_axes4.yaxis.set_ticks(np.arange(0,37,6))
main_axes4.xaxis.set_tick_params(labelsize=14)
main_axes4.yaxis.set_tick_params(labelsize=14)
main_axes4.set_xlabel('RMSD (nm)',fontsize=14,fontfamily='sans-serif',fontstyle='italic')
#fig.text(0.07,0.5,'Number of Rotatable Bonds',va='center',rotation='vertical',fontsize=18,fontfamily='sans-serif',fontstyle='italic')
#fig.text(0.5,0.05, 'RMSD (nm)',ha='center',fontsize=18,fontfamily='sans-serif',fontstyle='italic')
#
#plt.tight_layout()
plt.savefig('RMSD_figures/RMSD_rot_bonds_distribution.png',dpi=1000)