python科学绘图-histogram
春是生死的负重,夏是情欲的勃发,秋是因果的了断,冬是自我的修行
本篇博客延续了往期博客的风格,对我在Oligolysine和ATP形成liquid-liquid phase separation (LLPS)中用到的作图脚本进行了总结和汇总,方便有需要的朋友和自己下次使用。(这篇工作的发表也比较坎坷,chem. sci. 审稿了一个月,然后编辑给的重投,修改后再投后被拒并建议转投到PCCP后接受,也是第一次经历给机会修改后被毙掉的体验了。相关的Manuscript和Supporting information我作为附件一起放在了这篇博客后面,有兴趣的朋友可以看看。)
本教程中用到的相关库的版本如下,大家也可以不必局限于这个版本,本教程中的图都是正文和SI中出现的子图,整个图片的整合可以利用各自喜欢的软件来进行,我比较倾向于利用power point来进行组合,大家可以用Photoshop等工具来进行。
Requirement
- Python
3.8 - numpy
1.21.1 - pandas
1.3.1 - matplotlib
3.4.2
Figures
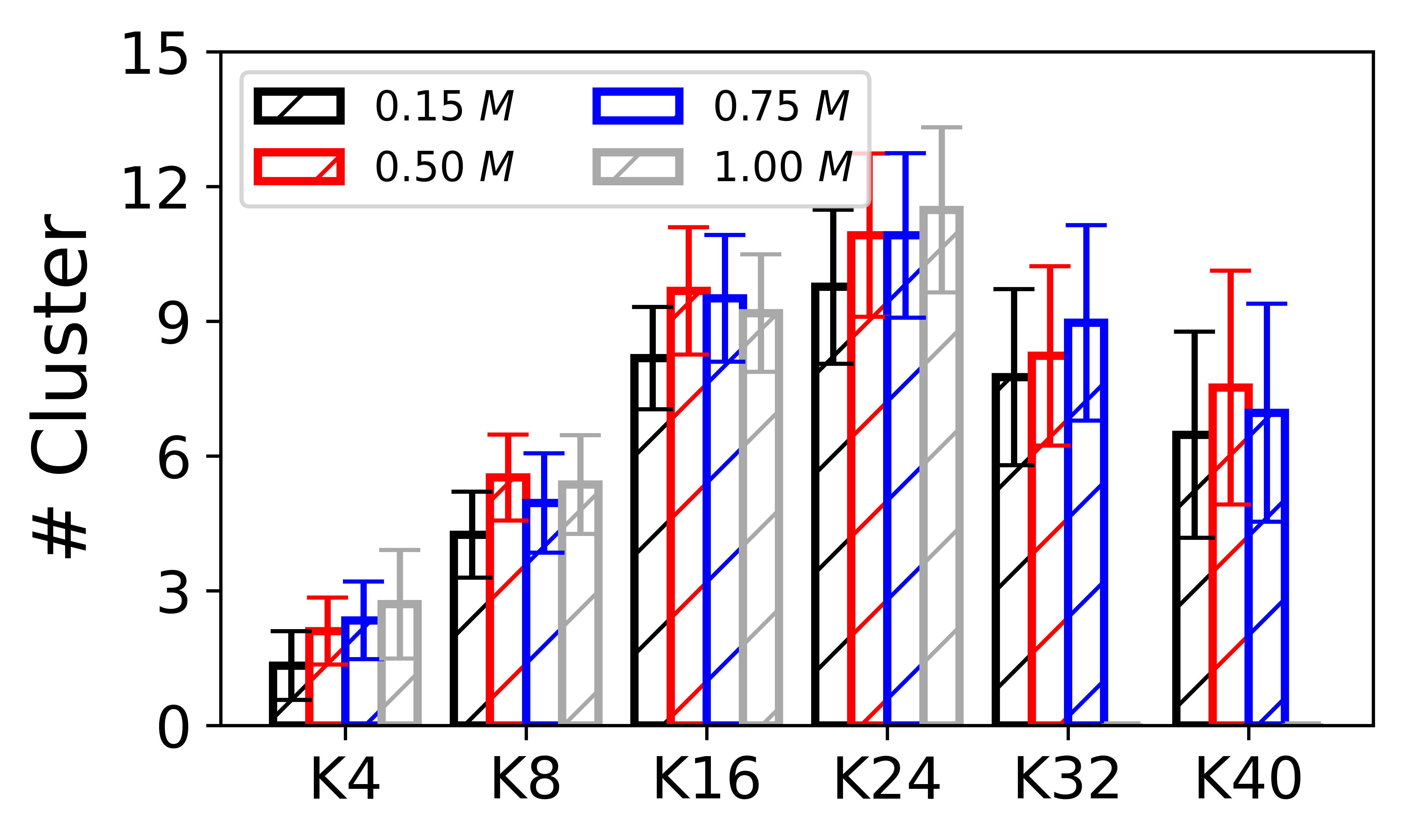
Cluster随Oligolysine长度的变化
highlighted points:
- 柱状图;
- 设置不同柱状图之间的间隔
#!/bin/python
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import os
# list for ionic concentrations utilized in this work
IONIC_STR = [0.15,0.50,0.75,1]
# function of loading data from files
def load_Kn(filename):
data = np.array([[0,0,0,0]])
if os.path.exists(filename):
data = np.loadtxt(filename)
else:
print('Not Exist!')
return data
# function of calculating avg, std values from data extracted from files
def get_clu(data):
results = {}
clu_num = data[:,1]
max_size = data[:,2]
ld_ratio = data[:,3]
num_mean, num_std = np.mean(clu_num), np.std(clu_num)
results['num_ave'] = num_mean
results['num_std'] = num_std
size_mean, size_std = np.mean(max_size), np.std(max_size)
results['size_ave'] = size_mean
results['size_std'] = size_std
ld_mean, ld_std = np.mean(ld_ratio), np.std(ld_ratio)
results['ld_ave'] = ld_mean
results['ld_std'] = ld_std
return results
# loading data for K4 (oligolysine of length 4)
results_k4_atp = {}
for ionic in IONIC_STR:
filename = 'K4/atp/{:.2f}M/cluster.dat'.format(ionic)
data_tmp = load_Kn(filename)
results_k4_atp[ionic] = data_tmp
# loading data for K8 (oligolysine of length 8)
results_k8_atp = {}
for ionic in IONIC_STR:
filename = 'K8/atp/{:.2f}M/cluster.dat'.format(ionic)
data_tmp = load_Kn(filename)
results_k8_atp[ionic] = data_tmp
# loading data for K16 (oligolysine of length 16)
results_k16_atp = {}
for ionic in IONIC_STR:
filename = 'K16/atp/{:.2f}M/cluster.dat'.format(ionic)
data_tmp = load_Kn(filename)
results_k16_atp[ionic] = data_tmp
# loading data for K24 (oligolysine of length 24)
results_k24_atp = {}
for ionic in IONIC_STR:
filename = 'K24/atp/{:.2f}M/cluster.dat'.format(ionic)
data_tmp = load_Kn(filename)
results_k24_atp[ionic] = data_tmp
# loading data for K32 (oligolysine of length 24)
results_k32_atp = {}
for ionic in IONIC_STR:
filename = 'K32/atp/{:.2f}M/cluster.dat'.format(ionic)
data_tmp = load_Kn(filename)
results_k32_atp[ionic] = data_tmp
# loading data for K40 (oligolysine of length 40)
results_k40_atp = {}
for ionic in IONIC_STR:
filename = 'K40/atp/{:.2f}M/cluster.dat'.format(ionic)
data_tmp = load_Kn(filename)
results_k40_atp[ionic] = data_tmp
# processing data
data_k4 ={}
for k,v in results_k4_atp.items():
#print(k,v)
data_k4[k]=get_clu(v)
data_k8 ={}
for k,v in results_k8_atp.items():
#print(k,v)
data_k8[k]=get_clu(v)
data_k16 ={}
for k,v in results_k16_atp.items():
#print(k,v)
data_k16[k]=get_clu(v)
data_k24 ={}
for k,v in results_k24_atp.items():
#print(k,v)
data_k24[k]=get_clu(v)
data_k32 ={}
for k,v in results_k32_atp.items():
#print(k,v)
data_k32[k]=get_clu(v)
data_k40 ={}
for k,v in results_k40_atp.items():
#print(k,v)
data_k40[k]=get_clu(v)
# plot 1: number of cluster as a function oligolysine length
x_ = ['K4','K8','K16','K24','K32','K40']
x = np.arange(len(x_))
width=0.4
fig,ax=plt.subplots(figsize=(5,3))
y_ave_015 = [data_k4[0.15]['num_ave'],data_k8[0.15]['num_ave'],data_k16[0.15]['num_ave'],data_k24[0.15]['num_ave'],data_k32[0.15]['num_ave'],data_k40[0.15]['num_ave']]
y_std_015 = [data_k4[0.15]['num_std'],data_k8[0.15]['num_std'],data_k16[0.15]['num_std'],data_k24[0.15]['num_std'],data_k32[0.15]['num_std'],data_k40[0.15]['num_std']]
y_ave_050 = [data_k4[0.50]['num_ave'],data_k8[0.50]['num_ave'],data_k16[0.50]['num_ave'],data_k24[0.50]['num_ave'],data_k32[0.50]['num_ave'],data_k40[0.50]['num_ave']]
y_std_050 = [data_k4[0.50]['num_std'],data_k8[0.50]['num_std'],data_k16[0.50]['num_std'],data_k24[0.50]['num_std'],data_k32[0.50]['num_std'],data_k40[0.50]['num_std']]
y_ave_075 = [data_k4[0.75]['num_ave'],data_k8[0.75]['num_ave'],data_k16[0.75]['num_ave'],data_k24[0.75]['num_ave'],data_k32[0.75]['num_ave'],data_k40[0.75]['num_ave']]
y_std_075 = [data_k4[0.75]['num_std'],data_k8[0.75]['num_std'],data_k16[0.75]['num_std'],data_k24[0.75]['num_std'],data_k32[0.75]['num_std'],data_k40[0.75]['num_std']]
y_ave_100 = [data_k4[1]['num_ave'],data_k8[1]['num_ave'],data_k16[1]['num_ave'],data_k24[1]['num_ave'],data_k32[1]['num_ave'],data_k40[1]['num_ave']]
y_std_100 = [data_k4[1]['num_std'],data_k8[1]['num_std'],data_k16[1]['num_std'],data_k24[1]['num_std'],data_k32[1]['num_std'],data_k40[1]['num_std']]
ax.bar(x-3*width/4, y_ave_015, yerr=y_std_015, width=width/2, color='white', edgecolor='black',hatch='/', ecolor='black', capsize=5,linewidth=2,label=r'$0.15~M$')
ax.bar(x-1*width/4, y_ave_050, yerr=y_std_050, width=width/2, color='white', edgecolor='red',hatch='/', ecolor='red', capsize=5,linewidth=2,label=r'$0.50~M$')
ax.bar(x+1*width/4, y_ave_075, yerr=y_std_075, width=width/2, color='white', edgecolor='blue',hatch='/', ecolor='blue', capsize=5,linewidth=2,label=r'$0.75~M$')
ax.bar(x+3*width/4, y_ave_100, yerr=y_std_100, width=width/2, color='white', edgecolor='darkgray',hatch='/', ecolor='darkgray', capsize=5,linewidth=2,label=r'$1.00~M$')
ax.legend(ncol=2)
ax.set_ylim(0,15)
ax.set_ylabel(r'# Cluster', fontsize=18,fontfamily='sans-serif')
ax.yaxis.set_ticks(np.arange(0,15.1,3))
ax.yaxis.set_tick_params(labelsize=14)
ax.xaxis.set_tick_params(labelsize=14)
plt.xticks(x,x_)
plt.savefig('231209_ionic_strength_cluster_num.png',dpi=1000,bbox_inches='tight')
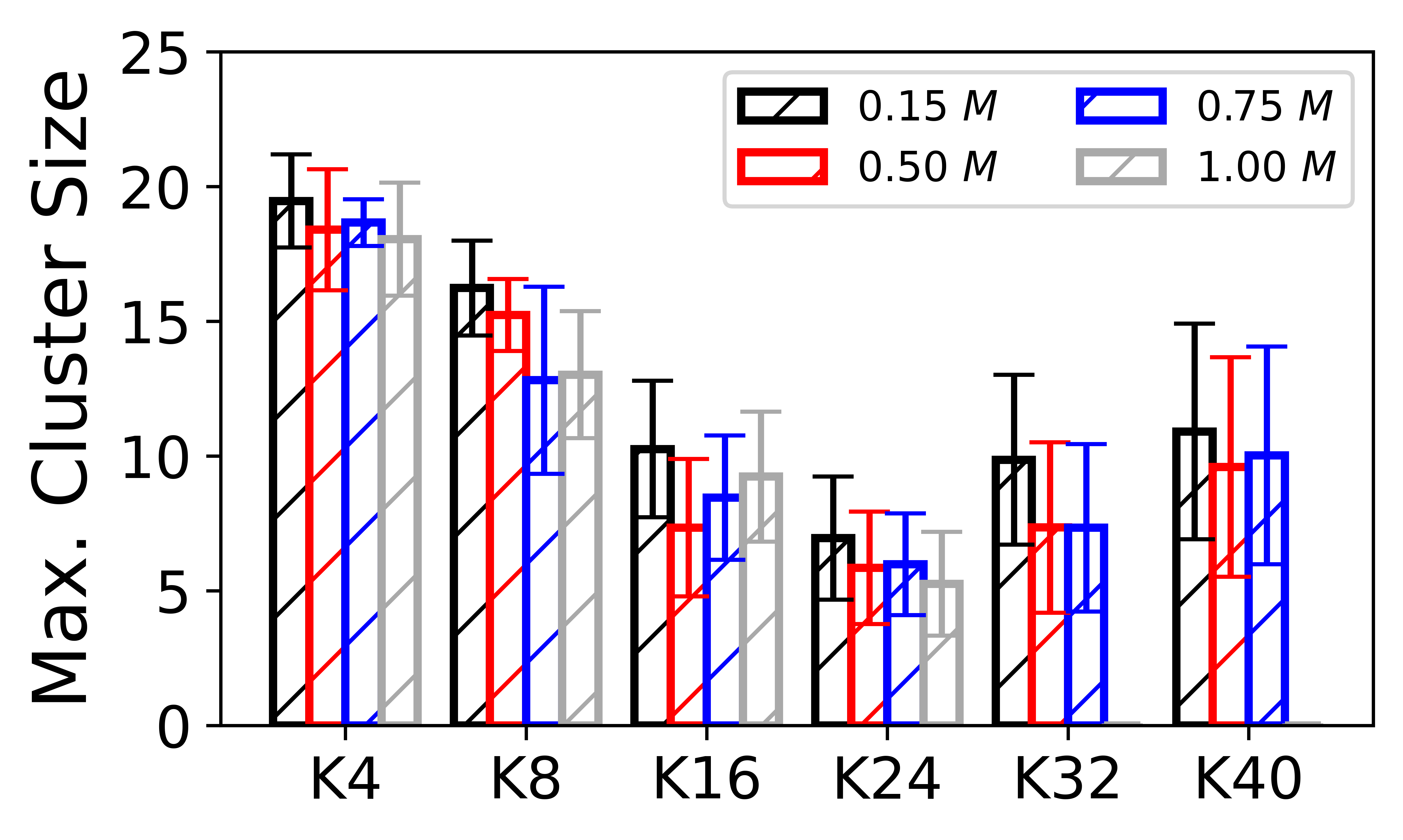
# plot 2: maximum size of cluster as a function oligolysine length
x_ = ['K4','K8','K16','K24','K32','K40']
x = np.arange(len(x_))
width=0.4
fig,ax=plt.subplots(figsize=(5,3))
y_ave_015 = [data_k4[0.15]['size_ave'],data_k8[0.15]['size_ave'],data_k16[0.15]['size_ave'],data_k24[0.15]['size_ave'],data_k32[0.15]['size_ave'],data_k40[0.15]['size_ave']]
y_std_015 = [data_k4[0.15]['size_std'],data_k8[0.15]['size_std'],data_k16[0.15]['size_std'],data_k24[0.15]['size_std'],data_k32[0.15]['size_std'],data_k40[0.15]['size_std']]
y_ave_050 = [data_k4[0.5]['size_ave'],data_k8[0.50]['size_ave'],data_k16[0.50]['size_ave'],data_k24[0.50]['size_ave'],data_k32[0.50]['size_ave'],data_k40[0.50]['size_ave']]
y_std_050 = [data_k4[0.5]['size_std'],data_k8[0.50]['size_std'],data_k16[0.50]['size_std'],data_k24[0.50]['size_std'],data_k32[0.50]['size_std'],data_k40[0.50]['size_std']]
y_ave_075 = [data_k4[0.75]['size_ave'],data_k8[0.75]['size_ave'],data_k16[0.75]['size_ave'],data_k24[0.75]['size_ave'],data_k32[0.75]['size_ave'],data_k40[0.75]['size_ave']]
y_std_075 = [data_k4[0.75]['size_std'],data_k8[0.75]['size_std'],data_k16[0.75]['size_std'],data_k24[0.75]['size_std'],data_k32[0.75]['size_std'],data_k40[0.75]['size_std']]
y_ave_100 = [data_k4[1]['size_ave'],data_k8[1]['size_ave'],data_k16[1]['size_ave'],data_k24[1]['size_ave'],data_k32[1]['size_ave'],data_k40[1]['size_ave']]
y_std_100 = [data_k4[1]['size_std'],data_k8[1]['size_std'],data_k16[1]['size_std'],data_k24[1]['size_std'],data_k32[1]['size_std'],data_k40[1]['size_std']]
ax.bar(x-3*width/4, y_ave_015, yerr=y_std_015, width=width/2, color='white', edgecolor='black',hatch='/', ecolor='black', capsize=5,linewidth=2,label=r'$0.15~M$')
ax.bar(x-1*width/4, y_ave_050, yerr=y_std_050, width=width/2, color='white', edgecolor='red',hatch='/', ecolor='red', capsize=5,linewidth=2,label=r'$0.50~M$')
ax.bar(x+1*width/4, y_ave_075, yerr=y_std_075, width=width/2, color='white', edgecolor='blue',hatch='/', ecolor='blue', capsize=5,linewidth=2,label=r'$0.75~M$')
ax.bar(x+3*width/4, y_ave_100, yerr=y_std_100, width=width/2, color='white', edgecolor='darkgray',hatch='/', ecolor='darkgray', capsize=5,linewidth=2,label=r'$1.00~M$')
ax.legend(ncol=2)
ax.set_ylim(0,25)
ax.set_ylabel(r'Max. Cluster Size', fontsize=18,fontfamily='sans-serif')
ax.yaxis.set_ticks(np.arange(0,25.1,5))
ax.yaxis.set_tick_params(labelsize=14)
ax.xaxis.set_tick_params(labelsize=14)
plt.xticks(x,x_)
plt.savefig('231209_ionic_strength_cluster_size.png',dpi=1000,bbox_inches='tight')
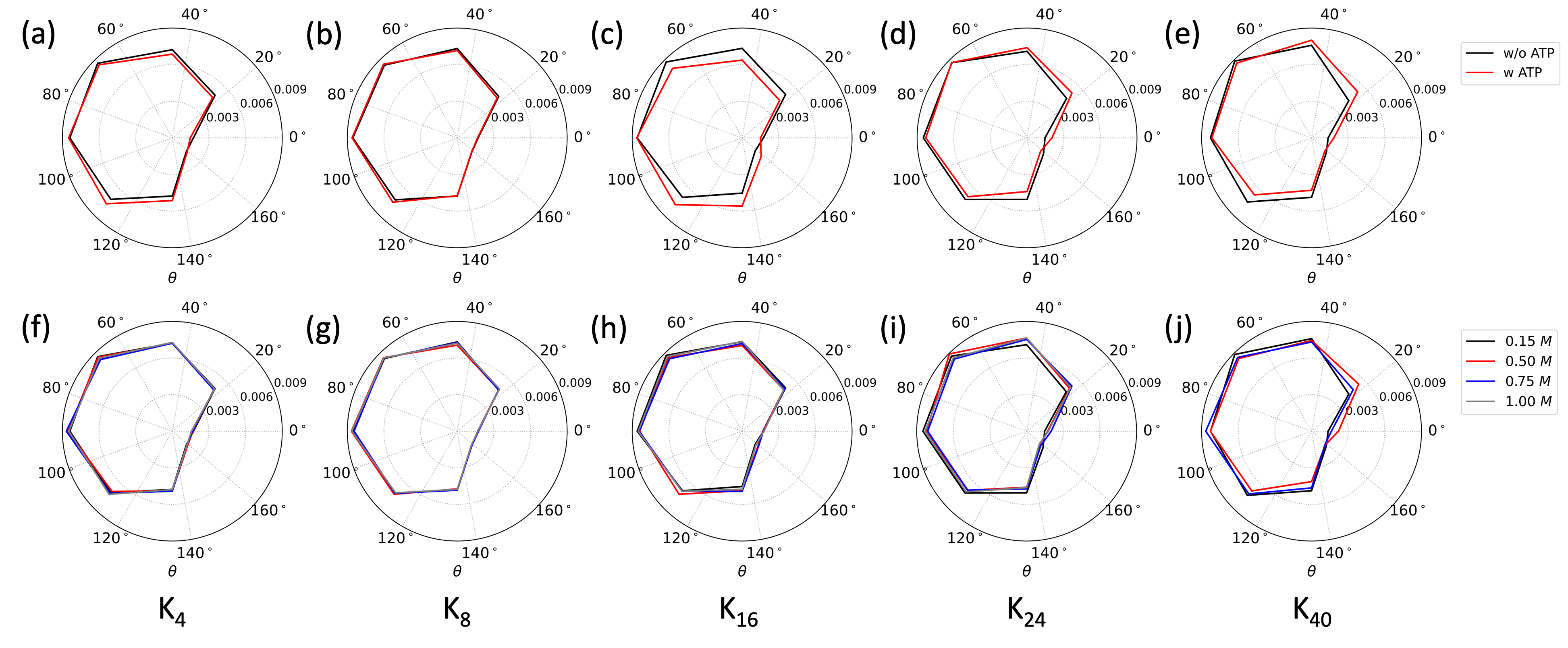
转动自由度随离子浓度的变化
highlighted points:
- radar plot;
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# function definition
def load_data(name:str,concs:list,has_atp:int)->dict:
results = {}
for conc in concs:
results[conc] = []
if has_atp!=0:
file = '{}/atp/{}M/dipole_orient.dat'.format(name,conc)
else:
file = '{}/no-atp/{}M/dipole_orient.dat'.format(name,conc)
data_tmp = np.loadtxt(file)
results[conc] = data_tmp
return results
# data processing
# w/o atp
concs = ['0.15','0.50','0.75','1.00']
concs_k40 = ['0.15','0.50','0.75']
namelist = ['k4','k8','k16','k24','k40']
results = {}
for name in namelist:
if name !='k40':
results[name] = load_data(name,concs,has_atp=0)
else:
results[name] = load_data(name,concs_k40,has_atp=0)
# data processing
# w atp
concs = ['0.15']
namelist = ['k4','k8','k16','k24','k40']
results_atp = {}
for name in namelist:
results_atp[name] = load_data(name,concs,has_atp=1)
# radar plot
ax = plt.subplot(projection='polar')
x_ = [r'$0^\circ$',r'$20^\circ$',r'$40^\circ$',r'$60^\circ$',r'$80^\circ$',r'$100^\circ$',r'$120^\circ$',r'$140^\circ$',r'$160^\circ$']
bins = 8
theta = np.linspace(0,2*np.pi,bins, endpoint=False)
theta = np.concatenate((theta,[theta[0]]))
hist_k4_015, bin_edges_k4_015 = np.histogram(results['k4']['0.15'][:,2],bins=bins,range=(0,180),density=True)
hist_k4_050, bin_edges_k4_050 = np.histogram(results['k4']['0.50'][:,2],bins=bins,range=(0,180),density=True)
hist_k4_075, bin_edges_k4_075 = np.histogram(results['k4']['0.75'][:,2],bins=bins,range=(0,180),density=True)
hist_k4_100, bin_edges_k4_100 = np.histogram(results['k4']['1.00'][:,2],bins=bins,range=(0,180),density=True)
hist_k4_015 = np.concatenate((hist_k4_015,[hist_k4_015[0]]))
hist_k4_050 = np.concatenate((hist_k4_050,[hist_k4_050[0]]))
hist_k4_075 = np.concatenate((hist_k4_075,[hist_k4_075[0]]))
hist_k4_100 = np.concatenate((hist_k4_100,[hist_k4_100[0]]))
ax.set_thetagrids(np.arange(0,360,40),x_)
ax.xaxis.set_tick_params(labelsize=16)
ax.yaxis.set_tick_params(labelsize=12)
ax.set_rlim(0,0.009)
ax.set_rgrids(np.arange(0.003,0.0091,0.003))
ax.plot(theta,hist_k4_015,'black',ms=5,label=r'$0.15~M$')
ax.plot(theta,hist_k4_050,'red',ms=5,label=r'$0.50~M$')
ax.plot(theta,hist_k4_075,'blue',ms=5,label=r'$0.75~M$')
ax.plot(theta,hist_k4_100,'grey',ms=5,label=r'$1.00~M$')
ax.set_xlabel(r'$\theta_{rot}$',fontsize=16)
#plt.legend(fontsize=14)
plt.grid(linestyle=':')
plt.tight_layout()
plt.savefig('no-atp_k4.png',dpi=1000,bbox_inches='tight')
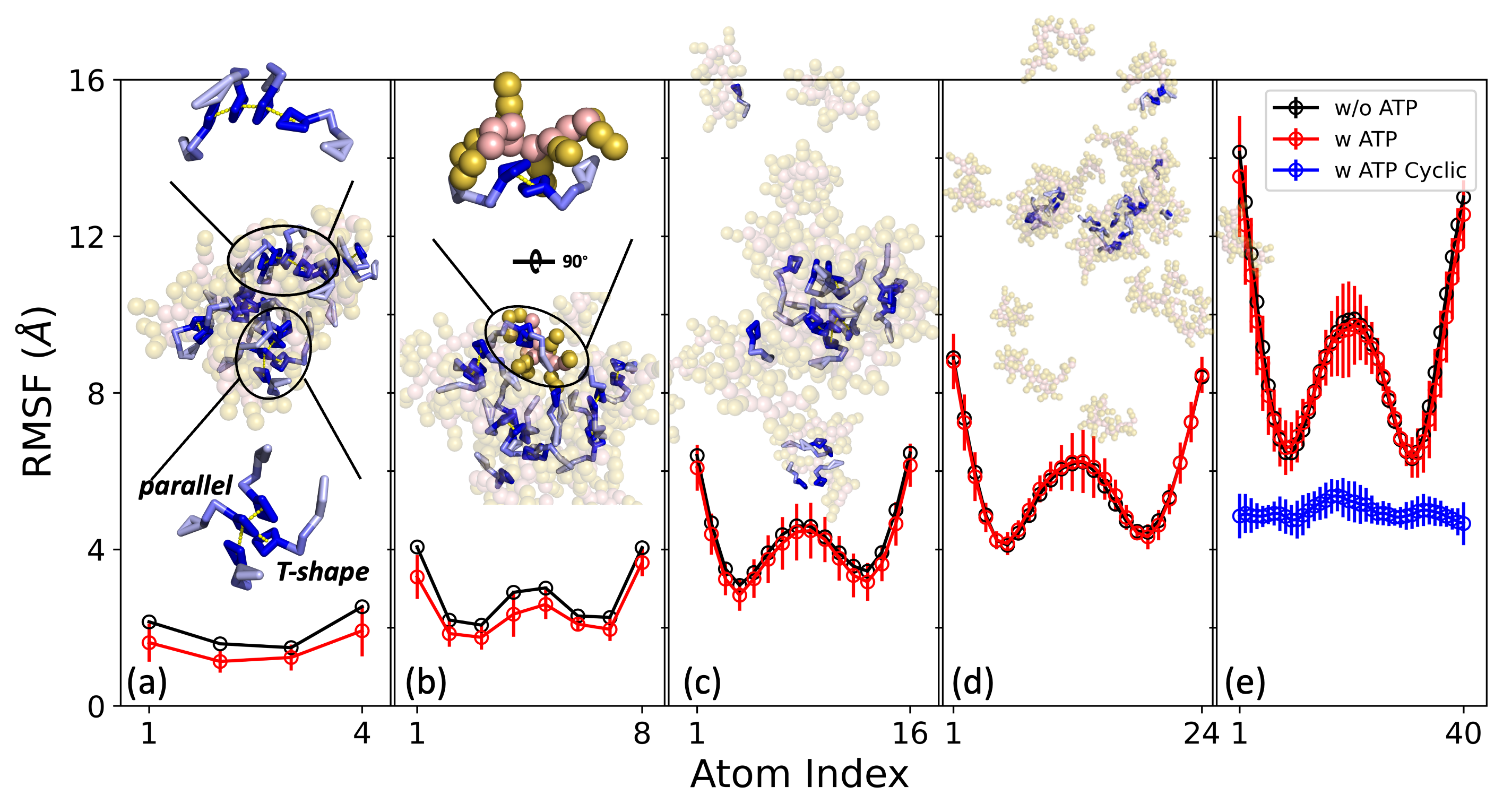
# 振动自由度的变化
highlighted points:
- 图片并列;
- 共享Y-axis
fig = plt.figure(figsize=(10,5))
ax0 = fig.add_subplot(1,5,1)
ax0.errorbar(np.arange(1,len(data_k4_noatp_015['idx'])+1), data_k4_noatp_015['mean'],yerr=data_k4_noatp_015['std'],mfc='none',marker='o',mec='black',ecolor='black',color='black',label='w/o ATP')
ax0.errorbar(np.arange(1,len(data_k4_atp_015['idx'])+1), data_k4_atp_015['mean'],yerr=data_k4_atp_015['std'],mfc='none',marker='o',mec='red',ecolor='red',color='red',label='w ATP')
ax0.set_ylabel(r'RMSF ($\AA$)', fontsize=18,fontfamily='sans-serif')
ax0.yaxis.set_tick_params(labelsize=14)
ax0.xaxis.set_tick_params(labelsize=14)
ax0.set_ylim(0,16)
ax0.set_xlim(0.6,4.4)
ax0.xaxis.set_ticks(np.arange(1,5,3))
ax0.yaxis.set_ticks(np.arange(0,16.1,4))
ax1 = fig.add_subplot(1,5,2)
ax1.errorbar(np.arange(1,len(data_k8_noatp_015['idx'])+1), data_k8_noatp_015['mean'],yerr=data_k8_noatp_015['std'],mfc='none',marker='o',mec='black',ecolor='black',color='black',label='w/o ATP')
ax1.errorbar(np.arange(1,len(data_k8_atp_015['idx'])+1), data_k8_atp_015['mean'],yerr=data_k8_atp_015['std'],mfc='none',marker='o',mec='red',ecolor='red',color='red',label='w ATP')
#ax1.set_ylabel(r'RMSF ($\AA$)', fontsize=18,fontfamily='sans-serif')
ax1.yaxis.set_tick_params(labelsize=14)
ax1.xaxis.set_tick_params(labelsize=14)
ax1.yaxis.set_minor_formatter(plt.NullFormatter())
ax1.yaxis.set_major_formatter(plt.NullFormatter())
ax1.set_ylim(0,16)
ax1.set_xlim(0.3,8.7)
ax1.xaxis.set_ticks(np.arange(1,9,7))
ax2 = fig.add_subplot(1,5,3)
ax2.errorbar(np.arange(1,len(data_k16_noatp_015['idx'])+1), data_k16_noatp_015['mean'],yerr=data_k16_noatp_015['std'],mfc='none',marker='o',mec='black',ecolor='black',color='black',label='w/o ATP')
ax2.errorbar(np.arange(1,len(data_k16_atp_015['idx'])+1), data_k16_atp_015['mean'],yerr=data_k16_atp_015['std'],mfc='none',marker='o',mec='red',ecolor='red',color='red',label='w ATP')
#ax1.set_ylabel(r'RMSF ($\AA$)', fontsize=18,fontfamily='sans-serif')
ax2.yaxis.set_tick_params(labelsize=14)
ax2.xaxis.set_tick_params(labelsize=14)
ax2.yaxis.set_minor_formatter(plt.NullFormatter())
ax2.yaxis.set_major_formatter(plt.NullFormatter())
ax2.set_ylim(0,16)
ax2.set_xlim(-1,18)
ax2.xaxis.set_ticks(np.arange(1,17,15))
ax3 = fig.add_subplot(1,5,4)
ax3.errorbar(np.arange(1,len(data_k24_noatp_015['idx'])+1), data_k24_noatp_015['mean'],yerr=data_k24_noatp_015['std'],mfc='none',marker='o',mec='black',ecolor='black',color='black',label='w/o ATP')
ax3.errorbar(np.arange(1,len(data_k24_atp_015['idx'])+1), data_k24_atp_015['mean'],yerr=data_k24_atp_015['std'],mfc='none',marker='o',mec='red',ecolor='red',color='red',label='w ATP')
#ax1.set_ylabel(r'RMSF ($\AA$)', fontsize=18,fontfamily='sans-serif')
ax3.yaxis.set_tick_params(labelsize=14)
ax3.xaxis.set_tick_params(labelsize=14)
ax3.yaxis.set_minor_formatter(plt.NullFormatter())
ax3.yaxis.set_major_formatter(plt.NullFormatter())
ax3.set_ylim(0,16)
ax3.set_xlim(0,25)
ax3.xaxis.set_ticks(np.arange(1,25,23))
ax4 = fig.add_subplot(1,5,5)
ax4.errorbar(np.arange(1,len(data_k40_noatp_015['idx'])+1), data_k40_noatp_015['mean'],yerr=data_k40_noatp_015['std'],mfc='none',marker='o',mec='black',ecolor='black',color='black',label='w/o ATP')
ax4.errorbar(np.arange(1,len(data_k40_atp_015['idx'])+1), data_k40_atp_015['mean'],yerr=data_k40_atp_015['std'],mfc='none',marker='o',mec='red',ecolor='red',color='red',label='w ATP')
ax4.errorbar(np.arange(1,len(data_k40_atp_015_cyc['idx'])+1),data_k40_atp_015_cyc['mean'],yerr=data_k40_atp_015_cyc['std'],mfc='none',marker='o',mec='blue',ecolor='blue',color='blue',label='w ATP Cyclic')
#ax1.set_ylabel(r'RMSF ($\AA$)', fontsize=18,fontfamily='sans-serif')
ax4.yaxis.set_tick_params(labelsize=14)
ax4.xaxis.set_tick_params(labelsize=14)
ax4.yaxis.set_minor_formatter(plt.NullFormatter())
ax4.yaxis.set_major_formatter(plt.NullFormatter())
ax4.set_ylim(0,16)
ax4.set_xlim(-3,44)
ax4.xaxis.set_ticks(np.arange(1,41,39))
ax4.legend(fontsize=10,ncol=1)
ax2.text(0.5,-1.8,'Atom Index',va='center',fontsize=18,fontfamily='sans-serif')
plt.tight_layout()
plt.subplots_adjust(hspace=0,wspace=0.015)
#plt.savefig('fluctuation_atp_effect.png',dpi=800,bbox_inches='tight')
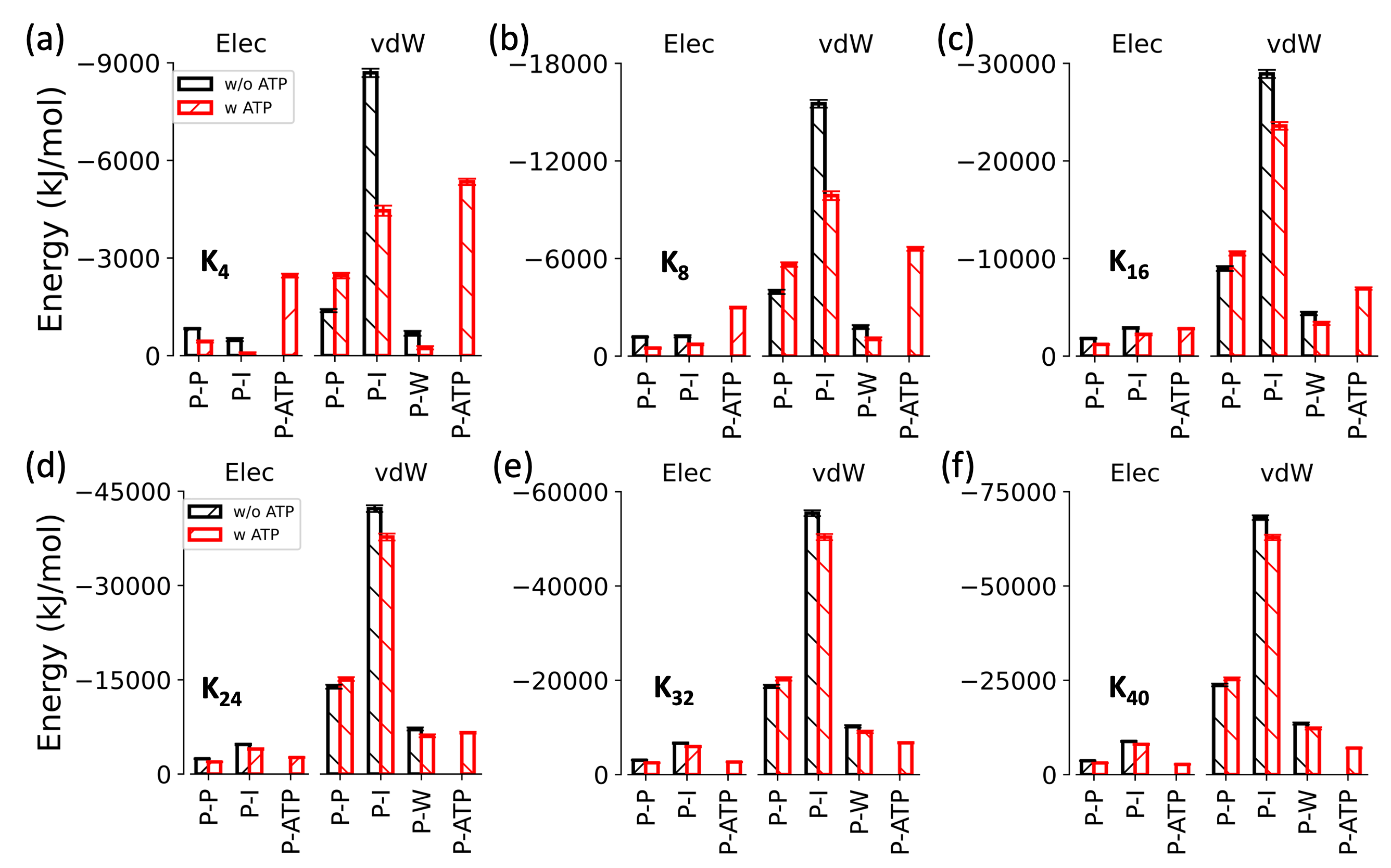
能量分解
highlighted points:
- 图片并列;
- 共享Y-axis同时隐藏Y-axis
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
def load_intE(filename,atp=0):
if atp:
results = {'pp-elec':[],'pp-lj':[],\
'pATP-elec':[],'pATP-lj':[],\
'pW-elec':[],'pW-lj':[],\
'pI-elec':[],'pI-lj':[]}
output = {'pp-elec':[],'pp-lj':[],\
'pATP-elec':[],'pATP-lj':[],\
'pW-elec':[],'pW-lj':[],\
'pI-elec':[],'pI-lj':[]}
with open(filename,'r') as f:
while True:
line = f.readline()
if not line:
break
else:
if line[:1] not in ['#','@']:
results['pp-elec'].append(float(line.split()[1]))
results['pp-lj'].append(float(line.split()[2]))
results['pATP-elec'].append(float(line.split()[3]))
results['pATP-lj'].append(float(line.split()[4]))
results['pW-elec'].append(float(line.split()[5]))
results['pW-lj'].append(float(line.split()[6]))
results['pI-elec'].append(float(line.split()[7]))
results['pI-lj'].append(float(line.split()[8]))
output['pp-elec'].append([np.mean(results['pp-elec']),np.std(results['pp-elec'])])
output['pp-lj'].append([np.mean(results['pp-lj']),np.std(results['pp-lj'])])
output['pATP-elec'].append([np.mean(results['pATP-elec']),np.std(results['pATP-elec'])])
output['pATP-lj'].append([np.mean(results['pATP-lj']),np.std(results['pATP-lj'])])
output['pW-elec'].append([np.mean(results['pW-elec']),np.std(results['pW-elec'])])
output['pW-lj'].append([np.mean(results['pW-lj']),np.std(results['pW-lj'])])
output['pI-elec'].append([np.mean(results['pI-elec']),np.std(results['pI-elec'])])
output['pI-lj'].append([np.mean(results['pI-lj']),np.std(results['pI-lj'])])
else:
results = {'pp-elec':[],'pp-lj':[],\
'pW-elec':[],'pW-lj':[],\
'pI-elec':[],'pI-lj':[]}
output = {'pp-elec':[],'pp-lj':[],\
'pW-elec':[],'pW-lj':[],\
'pI-elec':[],'pI-lj':[]}
with open(filename,'r') as f:
while True:
line = f.readline()
if not line:
break
else:
if line[:1] not in ['#','@']:
results['pp-elec'].append(float(line.split()[1]))
results['pp-lj'].append(float(line.split()[2]))
results['pW-elec'].append(float(line.split()[3]))
results['pW-lj'].append(float(line.split()[4]))
results['pI-elec'].append(float(line.split()[5]))
results['pI-lj'].append(float(line.split()[6]))
output['pp-elec'].append([np.mean(results['pp-elec']),np.std(results['pp-elec'])])
output['pp-lj'].append([np.mean(results['pp-lj']),np.std(results['pp-lj'])])
output['pW-elec'].append([np.mean(results['pW-elec']),np.std(results['pW-elec'])])
output['pW-lj'].append([np.mean(results['pW-lj']),np.std(results['pW-lj'])])
output['pI-elec'].append([np.mean(results['pI-elec']),np.std(results['pI-elec'])])
output['pI-lj'].append([np.mean(results['pI-lj']),np.std(results['pI-lj'])])
return output
# data processing
f_K4_noatp = 'K4/intE.xvg'
f_K4_atp = 'K4-ATP/intE.xvg'
f_K8_noatp = 'K8/intE.xvg'
f_K8_atp = 'K8-ATP/intE.xvg'
f_K16_noatp= 'K16/intE.xvg'
f_K16_atp = 'K16-ATP/intE.xvg'
f_K24_noatp= 'K24/intE.xvg'
f_K24_atp = 'K24-ATP/intE.xvg'
f_K32_noatp= 'K32/intE.xvg'
f_K32_atp = 'K32-ATP/intE.xvg'
f_K40_noatp= 'K40/intE.xvg'
f_K40_atp = 'K40-ATP/intE.xvg'
output_K4_noatp = load_intE(f_K4_noatp,atp=0)
output_K4_atp = load_intE(f_K4_atp,atp=1)
output_K8_noatp = load_intE(f_K8_noatp,atp=0)
output_K8_atp = load_intE(f_K8_atp,atp=1)
output_K16_noatp = load_intE(f_K16_noatp,atp=0)
output_K16_atp = load_intE(f_K16_atp,atp=1)
output_K24_noatp = load_intE(f_K24_noatp,atp=0)
output_K24_atp = load_intE(f_K24_atp,atp=1)
output_K32_noatp = load_intE(f_K32_noatp,atp=0)
output_K32_atp = load_intE(f_K32_atp,atp=1)
output_K40_noatp = load_intE(f_K40_noatp,atp=0)
output_K40_atp = load_intE(f_K40_atp,atp=1)
elec_k4_ave_noatp = [output_K4_noatp['pp-elec'][0][0],output_K4_noatp['pI-elec'][0][0]]
elec_k4_std_noatp = [output_K4_noatp['pp-elec'][0][1],output_K4_noatp['pI-elec'][0][1]]
lj_k4_ave_noatp = [output_K4_noatp['pp-lj'][0][0],output_K4_noatp['pW-lj'][0][0],output_K4_noatp['pI-lj'][0][0]]
lj_k4_std_noatp = [output_K4_noatp['pp-lj'][0][1],output_K4_noatp['pW-lj'][0][1],output_K4_noatp['pI-lj'][0][1]]
elec_k4_ave_atp = [output_K4_atp['pp-elec'][0][0],output_K4_atp['pI-elec'][0][0], output_K4_atp['pATP-elec'][0][0]]
elec_k4_std_atp = [output_K4_atp['pp-elec'][0][1],output_K4_atp['pI-elec'][0][1], output_K4_atp['pATP-elec'][0][1]]
lj_k4_ave_atp = [output_K4_atp['pp-lj'][0][0],output_K4_atp['pW-lj'][0][0],output_K4_atp['pI-lj'][0][0], output_K4_atp['pATP-lj'][0][0]]
lj_k4_std_atp = [output_K4_atp['pp-lj'][0][1],output_K4_atp['pW-lj'][0][1],output_K4_atp['pI-lj'][0][1], output_K4_atp['pATP-lj'][0][1]]
x_elec = ['P-P','P-I','P-ATP']
xelec = np.arange(len(x_elec))
x_lj = ['P-P','P-I','P-W','P-ATP']
xlj = np.arange(len(x_lj))
width=0.3
# figure plot
fig = plt.figure(figsize=(3,3))
#fig,ax = plt.subplots(figsize=(1.5,3))
Grid = plt.GridSpec(1,7,hspace=0.2,wspace=0.4)
ax01 = fig.add_subplot(Grid[0,:3])
ax02 = fig.add_subplot(Grid[0,3:],sharey=ax01)
ax01.bar(np.arange(len(x_elec)-1)-width/2, elec_k4_ave_noatp, yerr=elec_k4_std_noatp, width=width, color='white', edgecolor='black',hatch='/', ecolor='black', capsize=5,linewidth=2,label='w/o ATP')
ax01.bar(xelec+width/2, elec_k4_ave_atp, yerr=elec_k4_std_atp, width=width, color='white', edgecolor='red',hatch='/', ecolor='red', capsize=5,linewidth=2,label='w ATP')
ax02.bar(np.arange(len(x_lj)-1)-width/2, lj_k4_ave_noatp, yerr=lj_k4_std_noatp, width=width, color='white', edgecolor='black',hatch='\\', ecolor='black', capsize=5,linewidth=2,label='w/o ATP')
ax02.bar(xlj+width/2, lj_k4_ave_atp, yerr=lj_k4_std_atp, width=width, color='white', edgecolor='red',hatch='\\', ecolor='red', capsize=5,linewidth=2,label='w ATP')
#plt.xticks(xelec,x_elec,rotation='vertical')
ax02.tick_params(axis='y',which='both',left=False,labelleft=False)
ax02.spines['left'].set_visible(False)
ax02.spines['right'].set_visible(False)
ax02.spines['top'].set_visible(False)
ax02.yaxis.set_tick_params(labelsize=14)
ax02.xaxis.set_tick_params(labelsize=14,labelrotation=90)
#ax02.yaxis.set_major_locator([])
ax02.yaxis.set_ticks([])
ax02.xaxis.set_ticks(xlj)
ax02.set_xticklabels(x_lj)
ax02.set_title('vdW',fontsize=14)
ax01.set_ylim(0,-9000)
#ax2.set_ylim(0,12)
ax01.set_ylabel(r'Energy (kJ/mol)', fontsize=18,fontfamily='sans-serif')
ax01.yaxis.set_ticks(np.arange(0,-9000.1,-3000))
ax01.yaxis.set_tick_params(labelsize=14)
ax01.xaxis.set_tick_params(labelsize=14,labelrotation=90)
ax01.spines['right'].set_visible(False)
ax01.spines['top'].set_visible(False)
ax01.spines['left'].set_position(('data',-.6))
ax01.xaxis.set_ticks(xelec)
ax01.set_xticklabels(x_elec)
ax01.set_title('Elec',fontsize=14)
#ax02.set_yticks([])
ax01.legend(loc='upper right',fontsize=9)
#plt.tight_layout()
plt.savefig('K4_ene.png',dpi=1000,bbox_inches='tight')